paired end sequencing wikipedia
Since the average size of fragments in the library is 250 bp 50bp paired-end reads have been found to be optimum for Hi-C library sequencing. Paired-end sequencing facilitates detection of genomic rearrangements and repetitive sequence elements as well as gene fusions and novel transcripts.

What Are Paired End Reads The Sequencing Center
In addition to producing twice the number of reads for the same time and effort in library.
. In paired-end reading it starts at one read finishes this direction at the specified read length and then starts another round of reading from the opposite end of the fragment. Covaris and Sage - Complementary DNA Shearing and Size-selection Tools for Mate-pair Library Construction PAG 2012. Paired end sequencing refers to the fact that the fragments sequenced were sequenced from both ends and not just the one as was true for first generation sequencing.
Read 2 often called the reverse read extends from the Read 2 Adapter in the 5 3 direction. Since the beginning of 2013 this preparation has been based on Nextera technology. These reads are assumed to be identical to.
The larger inserts mate pairs can pair reads across greater distances. Paired end や mate pair という用語はどのようにライブラリが作られたかどうやってシーケンスされたかを示します どちらの手法も配列情報に加えてゲノム中における二つの read 間の物理的な距離の情報を与えます. For subsets of the assemblies we integrated these with additional supporting data to confirm and complement the synteny-based adjacencies.
Polony sequencing is generally performed on paired-end tags library that each molecule of DNA template is of 135 bp in length with two 1718 bp paired genomic tags separated and flanked by common sequences. For those not familiar with paired-end reads check out this post. Learn about the difference between Paired-End and Single-Run sequencing and why the former creates more precise alignments than the latter especiall.
There is a unique adapter sequence on both ends of the paired-end read labeled Read 1 Adapter and Read 2 Adapter. For your De novo genome assembly Fig. Expanded NGS Applications for the Pippin Prep DNA Size Selection System.
Identification of complex genomic rearrangements. The first sequencing step is started by targeting SP1 to generate the forward read. For the first test I took some sequence from the human genome hg19 and created two 100 bp reads from this region.
The preparation of mate pair libraries is designed to allow classical paired-end sequencing of both ends of a fragment with an original size of several kilobases. Therefore they are able to better cover highly. Digilab and Sage - Complementary DNA Shearing and Size-selection Tools for Mate-pair Library Construction AGBT 2011.
Pairs come from the ends of the same DNA strand. Combining data from mate pair sequencing with those from short-insert paired-end reads provides increased information for maximising sequencing coverage across a genome 1. Typical experimental design advice for expression analyses using RNA-seq generally assumes that single-end reads provide robust gene-level expression estimates in a cost-effective manner and that the additional benefits obtained from paired-end sequencing are not worth the additional cost.
The differences between PE and MP reads include. Paired-end sequencing allows users to sequence both ends of a fragment and generate high-quality alignable sequence data. Paired-end reading improves the ability to identify the relative positions of various reads in the genome making it much more effective than single-end reading in resolving structural rearrangements such as.
Six with physical mapping data that anchor scaffolds to chromosome locations 13 with paired-end RNA sequencing RNAseq data and three with new assemblies based on re-scaffolding or Pacific Biosciences. The current read length of this technique is 26 bases per amplicon and 13 bases per tag leaving a gap of 45 bases in each tag. Paired-end sequencing allows users to sequence both ends of a fragment and generate high-quality alignable sequence data.
Mate pair sequencing involves generating long-insert paired-end DNA libraries useful for a number of sequencing applications including. US7601499B2 US11448462 US44846206A US7601499B2 US 7601499 B2 US7601499 B2 US 7601499B2 US 44846206 A US44846206 A US 44846206A US 7601499 B2 US7601499 B2 US 7601499B2 Authority US United States Prior art keywords nucleic acid target nucleic adaptor dna. Read 1 often called the forward read extends from the Read 1 Adapter in the 5 3 direction towards Read 2 along the forward DNA strand.
Thirty-six or 50 bp reads are sufficient to identify most chromatin interacting pairs using Illumina paired-end sequencing. Paired-end sequencing facilitates detection of genomic. Typical experimental design advice for expression analyses using RNA-seq generally assumes that single-end reads provide robust gene-level expression estimates in a.
It has very nice and simple illustrations along with explanations on the terminology used in paired-end sequencing. In conventional paired-end sequencing you simply sequence using the adapter for one end and then once youre done you start over sequencing using the adapter for the other end. This means your two reads are the reverse complement of the 100 3-most bases of the Watson strand and the Crick strand.
This can be very helpful e. As with everything you get what you pay for- paired end sequencing will always be the better option but for differential expression analysis it is likely not worth the additional cost. However in many cases eg with Illumina NextSeq and NovaSeq.
The figure shows the workflow for mate-pair library preparation for Illumina sequencing. Library preparation protocols -- In short PE protocols attach an adapter SP1 to the fwd end and another adapter SP2 to the reverse end. Paired end sequencing Download PDF Info Publication number US7601499B2.
Now lets get started.

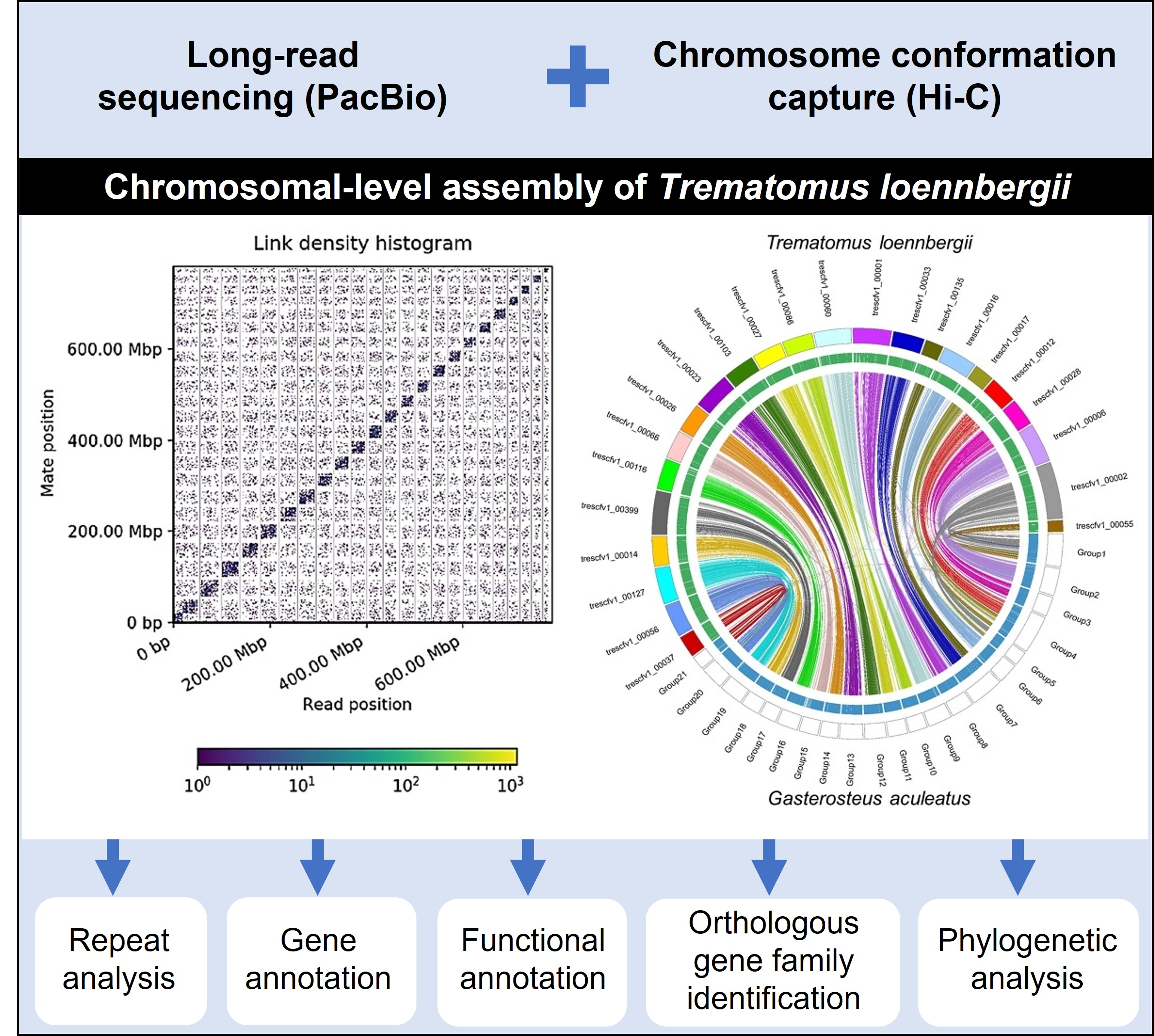
Diversity Free Full Text Chromosomal Level Assembly Of Antarctic Scaly Rockcod Trematomus Loennbergii Genome Using Long Read Sequencing And Chromosome Conformation Capture Hi C Technologies Html

What Are Paired End Reads The Sequencing Center

Paired End Shotgun Sequencing Okinawa Institute Of Science And Technology Graduate University Oist

What Is Mate Pair Sequencing For
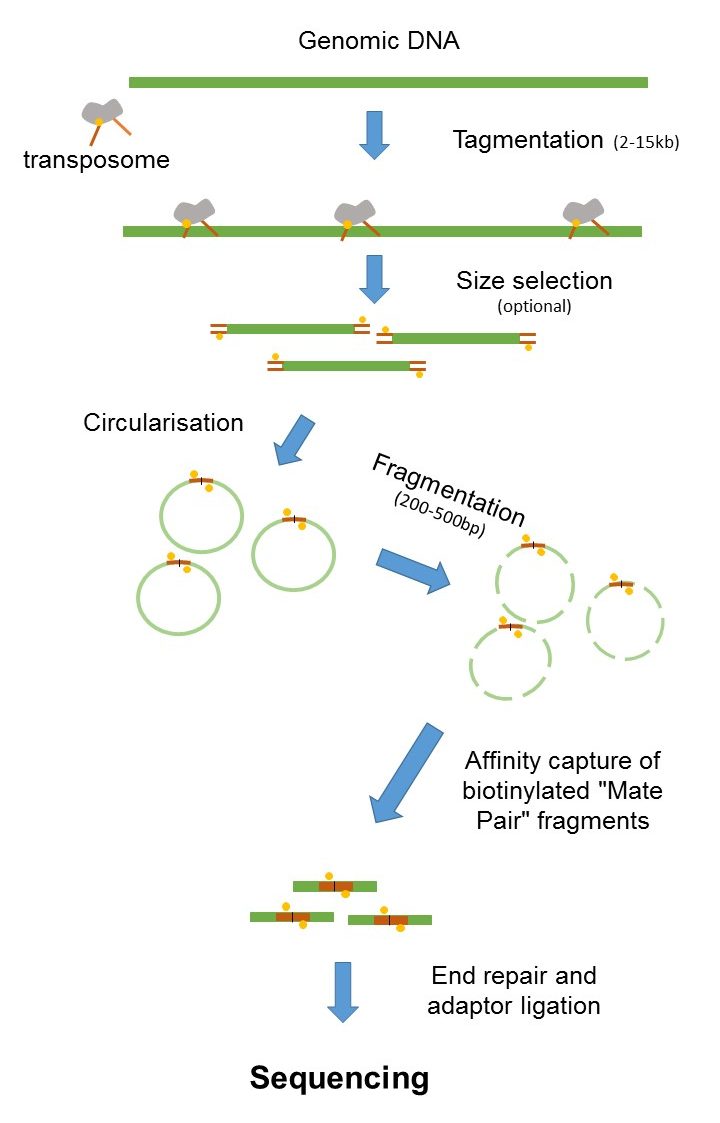
Mate Pair Sequencing France Genomique

What Is The Difference Between Single End And Paired End In Sequencing

What Is Mate Pair Sequencing For

Bacterial Chromosome Replication Biology Lessons Chromosome Biology

Bacterial Chromosome Replication Biology Lessons Chromosome Biology
Difference Between Whole Genome Sequencing And Next Generation Sequencing Difference Between

File Chromosome Conformation Techniques Jpg Wikipedia
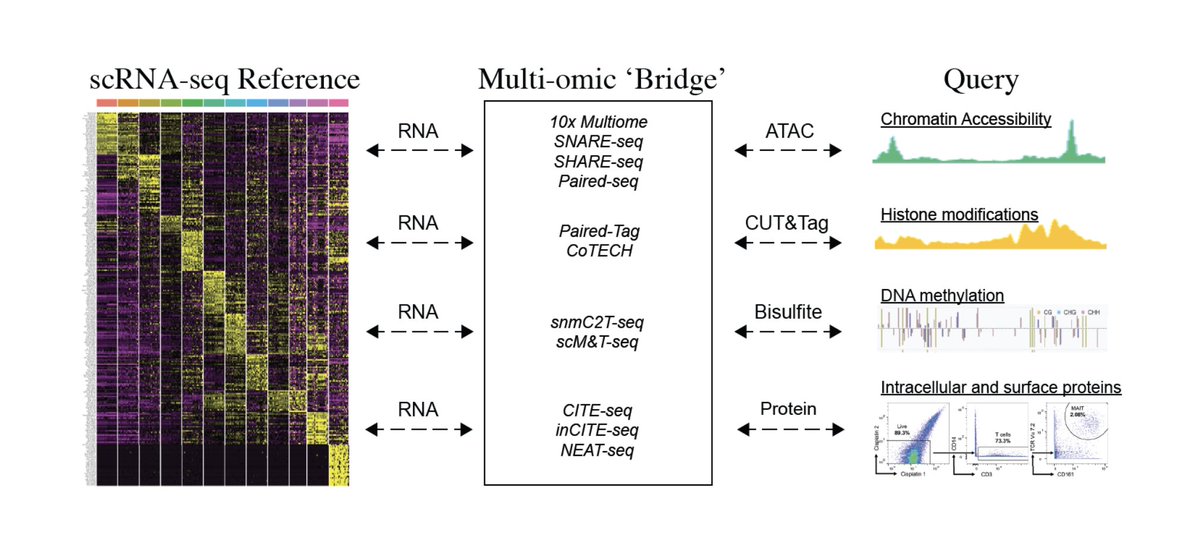
Rahul Satija Satijalab Twitter
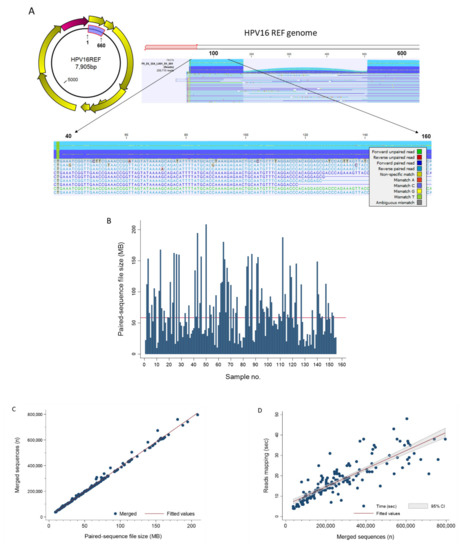
Pathogens Free Full Text Hpv Deepseq An Ultra Fast Method Of Ngs Data Analysis And Visualization Using Automated Workflows And A Customized Papillomavirus Database In Clc Genomics Workbench Html
Plos Computational Biology Informatics For Rna Sequencing A Web Resource For Analysis On The Cloud




